Hey everyone,
I had the pleasure of attending Gurobi’s Energy Innovation Summit last week, where I was very impressed with the contribution of Dr. Hofmann and Dr. Frysztacki from OET which showed their dramatic speed improvements for PyPSA when switiching over from Pyomo to Linopy.
Potentially already in the works? Any hindrances that I as a relatively “surface-level” oemof user would be unaware of?
Cheers,
Mo
Hi @moritz_reuter,
I think a transition would create considerable effort. Without knowing the details, I cannot tell if it will be worth it. In particular, Linopy highlights “N-D labeled variables”, which sounds like something we already apply in Pyomo, but many others do not. I do not have full 1:1 comparisons but only know one of oemof.solph and OSeMOSYS. Here is one example table:
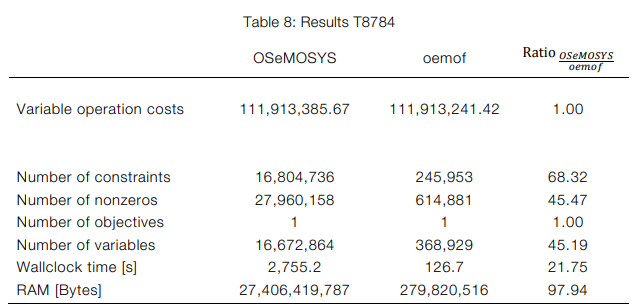
Source: FULLTEXT01.pdf (diva-portal.org)
Honestly, I think a clean and comprehensible API would be more appealing than an (unclear) performance improvement. But that also will just happen if someone can contribute time to at least implement a benchmark case.
Cheers,
Patrik
Hi @pschoen,
I would say a significant performance boost is guaranteed by our experience!
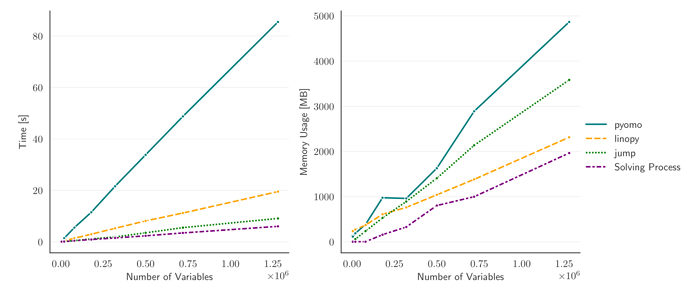
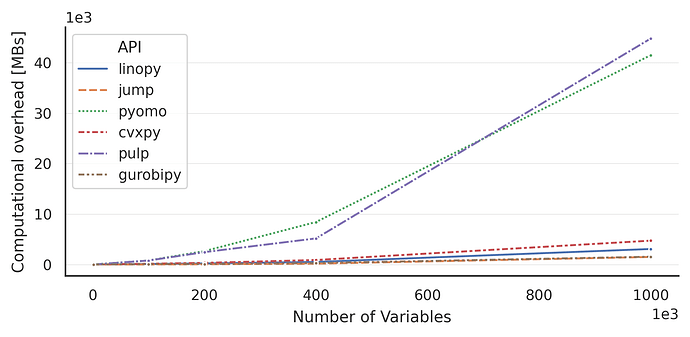
Here are some benchmarks compared with pyomo and other Julia-based tools:
https://linopy.readthedocs.io/en/latest/benchmark.html
Switching to linopy with PyPSA has meant plainly that we can solve 2-3 times larger problems without extra resources, and even solve models we couldn’t solve before in reasonable time.
Almost all constraints in capacity expansion models will be “N-D labeled”, and it’s not a problem if not!
I don’t think the solver interface backend would impact the API. In PyPSA we have added a switch for the user to choose which solver interface they’d like to use.
The comparison in the table I could not quite understand. It seems that two different optimisation problems are compared?
Best wishes,
Fabian
I agree, I’d also consider API and backend independent.
It’s the same problem formulated using two different tools, namely OSeMOSYS and (oemof.) solph. OSeMOSYS creates one variable per component, solph creates one n-dimensional variable per component type, where the individual components are an index.
So, I believe that we currently have a performance that is pretty good compared to other Pyomo based tools, thus the benefit of switching might (or might not) be smaller. Taking into account the effort for the transition, I won’t push the topic too much if there is no benchmark showing some parts of solph being translated to Linopy.